
The Eco'n'OMICs project
Pesticides, biocides or pharmaceuticals can display adverse effects in non-target organisms. This may threaten populations with far-reaching consequences for the ecosystem. Therefore, European legislation requires manufacturers to provide data for environmental risk assessment of active substances for registration. The commonly applied OECD tests are time and costconsuming and utilize a substantial number of animals. Thus, they are conducted only in the final stage of substance development, bearing the risk of failing registration due to adverse environmental effects. The Eco'n'OMICs project aims at an early ecotoxicological risk prediction for development compound classes based on their induced molecular changes in aquatic model organisms. We apply OMICs to identify gene expression changes as molecular biomarkers for a number of ecotoxicologically well characterized model substances covering a broad range of modes-of-action (MoA). The MoA-specific molecular signatures are linked with the corresponding adverse environmental effects and with the substance structures. The resulting data base is used to develop targeted molecular and in silico screening approaches in order to rank members of development compound classes based on their ecotoxic potential.
Background
After uptake by environmental organisms ecotoxic compounds induce molecular events, which initiate a cascade of processes across different organizational layers finally resulting in adverse effects at the level of the organism and the population. A direct link between such molecular initiating events (MIEs) and the subsequent adverse effects is described by the Adverse Outcome Pathway (AOP) concept. As an early response to MIEs, the organism reacts by highly specific changes in gene expression and cell metabolism. That way, the active substance of a pharmaceutical with a given MoA can for example induce gene expression changes in aquatic organisms. This can lead to liver toxicity, neurotoxicity or a reduced reproductive capacity at the organism level and therefore may affect the population. In contrast, non-hazardous active substances do not induce MIEs in environmental organisms and thus do not cause adverse effects at the molecular or organism levels. Hence, highly specific changes in gene expression precede every specific adverse environmental effect. These changes can be assessed in order to obtain an early prediction of ecotoxicity. Novel molecular biology and bioinformatics methods, referred to as OMICs analyses, allow for a sensitive and global detection of gene expression changes in a wide range of organisms. For several years, OMICs have been applied to assess efficacy and side effects of pharmaceutical active substances in humans. Only recently have OMICs started to be used for the identification of environmental side effects of pesticides, biocides and pharmaceuticals, consequently establishing the novel field of "ecotoxicogenomics". This development opens up new avenues for the detection of early molecular fingerprints and biomarkers in order to identify and differentiate MoAs of ecotoxic substances.
Approach
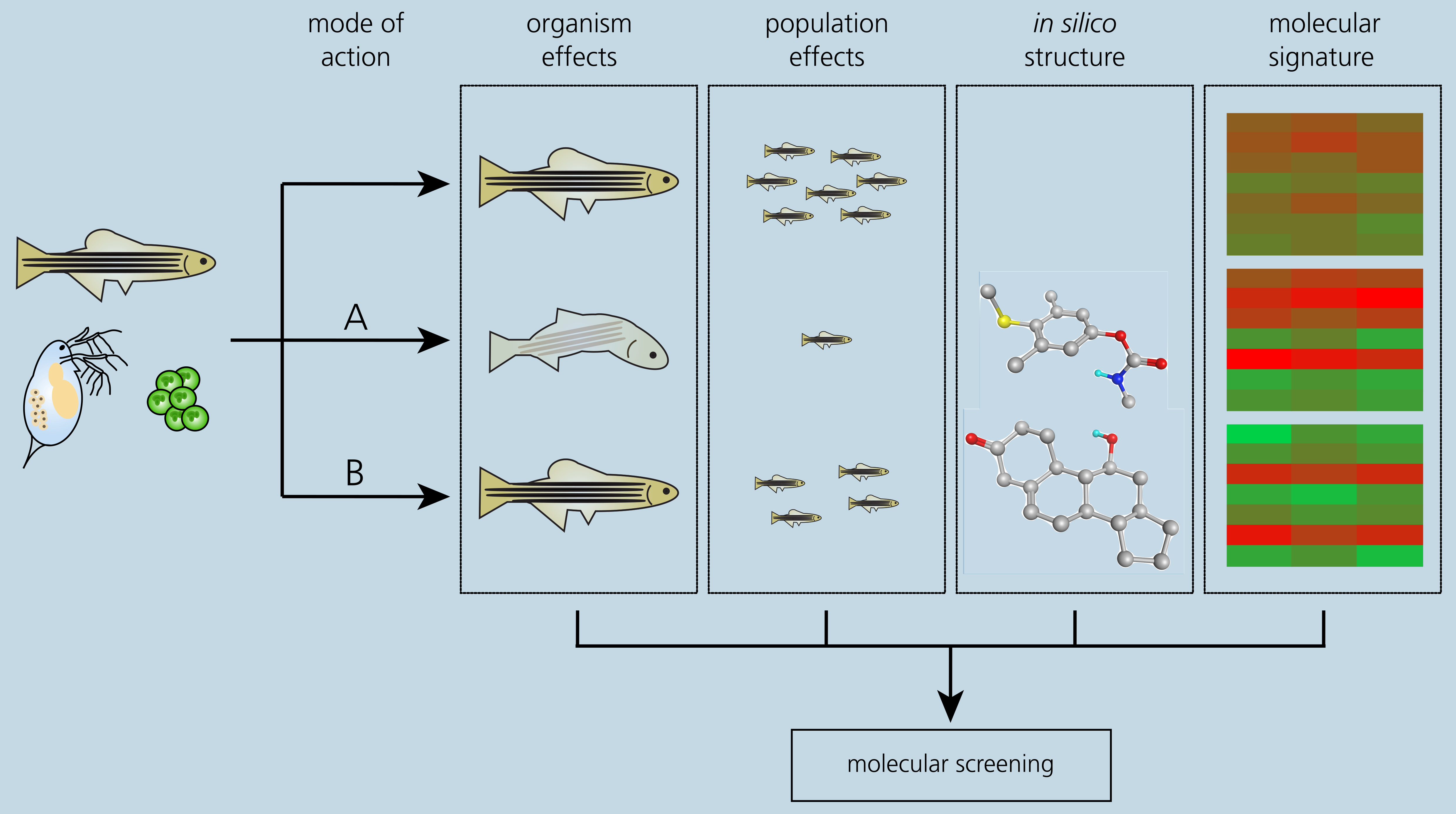
We combine recent transcriptomic and proteomic techniques with ecotoxicological approaches to generate a data base linking substance-specific gene expression signatures with adverse effects on the organism and the population. Therefore, aquatic model organisms such as fish larvae, water flea or algae are exposed to sublethal concentrations of reference substances in an environment based on standard OECD test guidelines. The reference substances are selected to be ecotoxicologically well characterized chemicals with known adverse effects, covering a broad range of MoAs. Our data base covers the substance's MoA, the compound structure, adverse effects on the organism and the population as well as the OMICs-generated molecular fingerprint in aquatic non-target organisms (Figure 4). These data represent the basis for the development of two screening pipelines to rank members of development compound classes based on their ecotoxic potential:
- A structure-activity-relationship-based in silico approach to predict MoAs and potential adverse effects
- A targeted OMICs approach utilizing the Eco'n'OMICs molecular fingerprint data base to predict the ecotoxic potential of development compounds.
Both approaches aim at providing an early identification of active substance candidates with low ecotoxic potential. Hence, they will support a time and costefficient development of environmentally friendly substances.
 Fraunhofer-Institut für Molekularbiologie und Angewandte Oekologie IME
Fraunhofer-Institut für Molekularbiologie und Angewandte Oekologie IME