Ecotoxicogenomic Screening for Ecotoxicological Risk Prediction


The active ingredients of pesticides, biocides, pharmaceuticals or cosmetics enter the environment either deliberately or through their use on humans or materials, where they can have a harmful effect on non-target organisms. Testing the environmental impact of an ever-increasing number of newly developed active ingredients represents an ever-increasing cost factor for the active ingredient-producing industry. In addition, the OECD tests required by law are time-consuming and resource-intensive and thus not compatible with the screening of substance precursors.
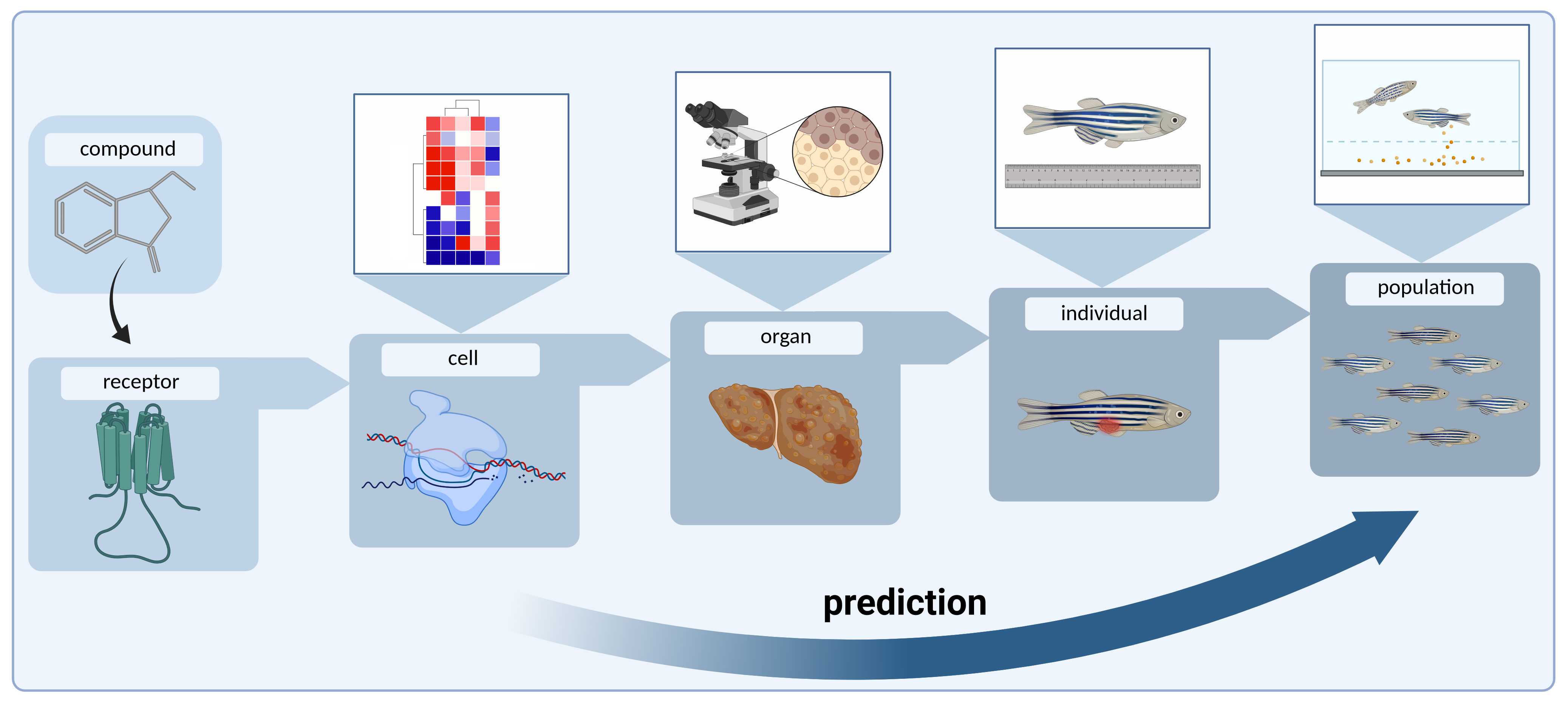
The application of state-of-the-art OMICs methods enables the sensitive and simultaneous genome-wide identification of substance-induced molecular changes in ecologically relevant organisms at the DNA (epigenome), RNA (transcriptome) and protein (proteome) levels. Knowledge of the molecular changes caused by a substance and their direct link to phenotypes and population effects opens up the possibility of early identification of ecotoxic substances and facilitates the classification of substances in terms of their environmental impact. Furthermore, such systems biology investigations can be carried out with minimal amounts of bioresources, making them a promising approach for the early detection of the environmental impact of substances as part of a screening process. At the same time, they enable a significant reduction in the number of animal experiments.
The OMICs-based identification of such pathways, which are disrupted by a substance, contributes significantly to the establishment of Adverse Outcome Pathways (AOPs). The AOP concept is used to directly link initial molecular events with the resulting adverse effects for the organism and the population and is also receiving increasing attention in regulatory substance assessment.
The Ecotoxicogenomics department combines ecotoxicological guideline testing with OMICs methods (RNA-Seq, quantitative LC-MS/MS) to detect early molecular changes that precede adverse organismic and population effects. Our molecular fingerprint database is used in screening methods to predict the environmental risks of substance precursors. The availability of such environmental side effect screening during industrial drug development avoids the costly further development of precursors with a high ecotoxic potential and enables the sustainable development of environmentally safe active ingredients.