Forisomes: multi-talents of biotechnology
Forisomes are unique, structural multiprotein complexes occurring exclusively in the phloem of plants belonging to the legume family. When the tissue is injured, they undergo a calcium-induced, reversible and ATP-independent structural change from a spindle-shaped to a plug-like form. In nature, this serves to rapidly close wounds within milliseconds and prevents the loss of the nutrient-rich phloem sap.
Using modern molecular biology methods, we are investigating the structure and reaction principle of these mechanoproteins. We succeeded in knowledge-based production of large quantities of functional, artificial forisomes and demonstrated their suitability as smart biomaterials.
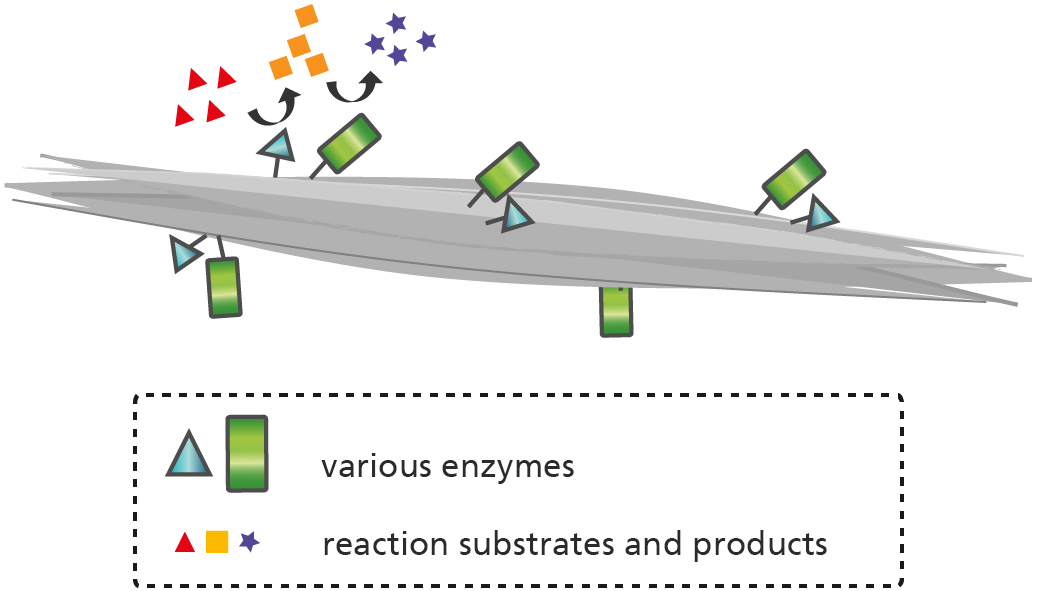
The fusion of forisomes with enzymes, so-called "forizymes", enables the stepwise production and immobilization of single and multi-enzyme complexes, whose special modification allows precise functionalization in microchannels of lab-on-a-chip systems. The combination of their catalytic and stimulus-dependent activity opens up new attractive fields of application in biosensors and microfluidics.
 Fraunhofer Institute for Molecular Biology and Applied Ecology IME
Fraunhofer Institute for Molecular Biology and Applied Ecology IME