The acceleration and improvement of plant breeding is necessary to meet the demands of the growing world population, especially as the amount of available farmland declines and crop failure becomes more frequent due to the extreme weather caused by climate change. New and improved crop varieties must be developed more quickly than before to prevent future food shortages. Without transgenic approaches, this can be only achieved by better and faster plant breeding techniques. Plant breeding requires innovative analytical techniques that allow the genetic properties of young seedlings to be identified long before the corresponding phenotypes appear.
Current molecular breeding approaches such as selection with markers and advanced reproductive technologies (SMART) or marker assisted selection (MAS) can improve breeding to a certain degree, but they also have disadvantages which prevent them making the breakthrough needed for the rapid generation of plants with multiple desirable properties.
Fraunhofer IME therefore aims to develop novel technological approaches that allow the rapid and high-throughput analysis of traits at the molecular level so that the selection process can be accelerated.
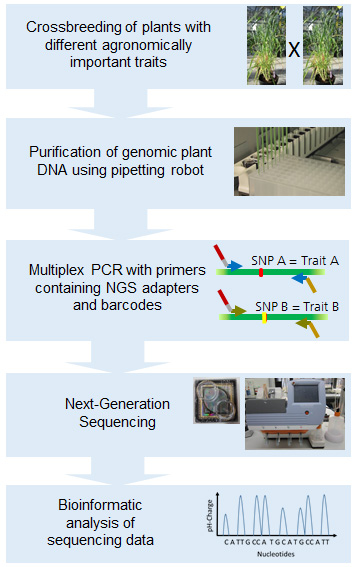
We have recently developed several new approaches for the analysis of plant breeding, including a multiplex PCR approach to determine zygosity and allelic distribution in maize, as well as an innovative next-generation sequencing approach for the high-throughput selection of plants with specific desirable traits. The next-generation sequencing of short PCR products spanning or flanking polymorphisms of interest in major crops such as barley and maize can provide robust genotyping data for the rapid determination of genotype and zygosity. This method can be applied to large panels of plants because up to 80 million individual reads can be generated and analyzed in one sequencing run. Hundreds of samples from different lines and/or traits can therefore be pooled and analyzed in parallel. These findings are significant because plant breeders may need to screen large populations for multiple traits to identify elite individuals. Our next-generation sequencing method therefore provides a simple and inexpensive approach for the rapid and accurate genotyping of natural polymorphisms, which can be applied in many economically relevant crops to achieve significantly faster and more accurate breeding.
 Fraunhofer Institute for Molecular Biology and Applied Ecology IME
Fraunhofer Institute for Molecular Biology and Applied Ecology IME