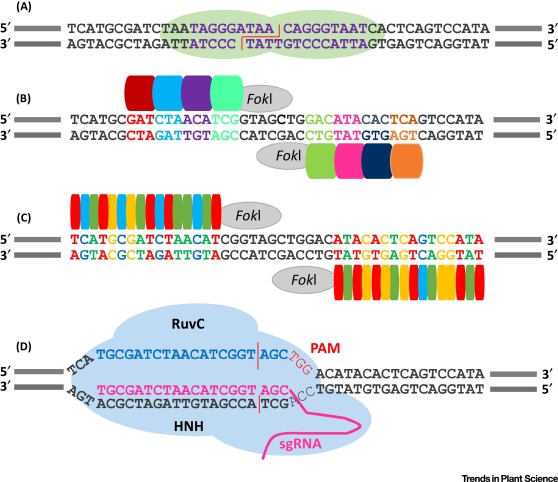
Site-specific nucleases introduce double-strand breaks at pre-determined DNA sequences, allowing precise and efficient genome engineering. Various targeted modifications can be generated by site-specific nucleases, including simple gene knockouts, complex gene conversions that require precise nucleotide exchange, transgene insertion, and molecular trait stacking. Genome editing can therefore accelerate plant breeding by facilitating the precise and predictable modification of endogenous genes directly in an elite background as well as the introduction of completely new traits, even complete biosynthesis pathways, in a controlled manner.
We use both zinc finger nucleases and the CRISPR/Cas9 system in plants and plant cell cultures to perform single and multiple gene knockouts, and to understand and address the challenges raised by the targeted integration of large transgene fragments.
 Fraunhofer Institute for Molecular Biology and Applied Ecology IME
Fraunhofer Institute for Molecular Biology and Applied Ecology IME