Proteome analysis allows the investigation of dynamic protein profiles in complex biological systems and is often combined with other “omic” techniques to describe and understand biochemical and physiological processes in cells, tissues and whole organisms. Changes in the proteome reveal how proteins interact to influence cellular processes, and show how the proteins themselves are affected by the environment.
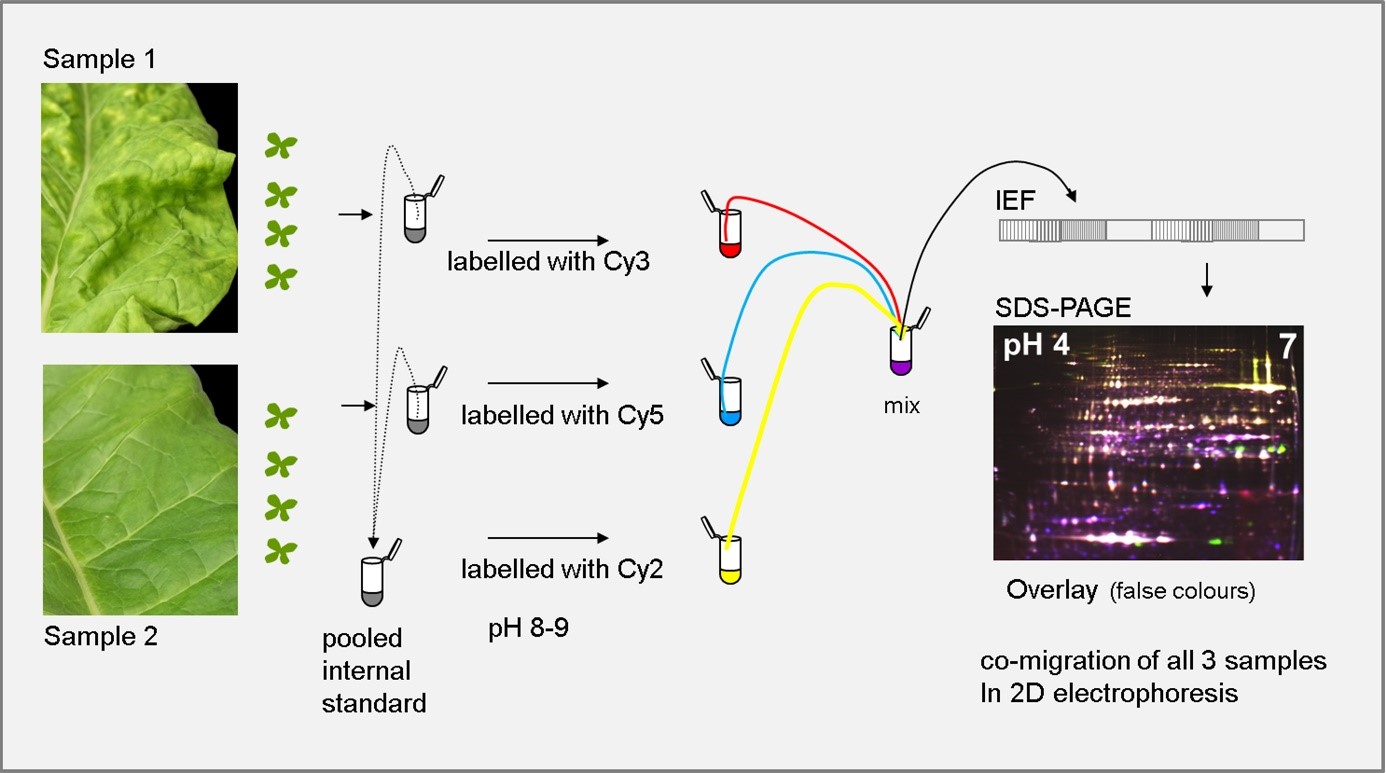
At Fraunhofer IME, proteome analysis is carried out by two-dimensional differential in-gel electrophoresis (2D-DIGE). This technique allows us to compare and accurately quantify the differences among several different samples simultaneously (e.g. comparison of different time points) using different fluorescent labels. Protein spots showing changes in abundance are analyzed by electrospray ionization ion trap mass spectrometry (ESI-IT-MS). Our 2D-DIGE and ESI-IT-MS data are processed using the DeCyder, Mascot and Compass software packages to identify the key proteins in each set of samples.
 Fraunhofer Institute for Molecular Biology and Applied Ecology IME
Fraunhofer Institute for Molecular Biology and Applied Ecology IME