Initial situation and project objective
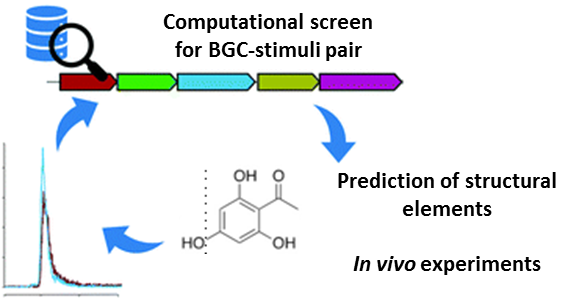
Microbial natural products (NPs) have always been a major inspiration for development of anti-infectives and control agents. Considered as none-essential metabolites for vegetative growth and reproduction of a microbe, but rather providing an evolutionary fitness advantages to changing environmental conditions, genes for NP biosynthesis are not constitutively expressed under laboratory conditions. This phenomenon is a constant challenge in cultivation-dependent but also -independent NP discovery campaigns demanding for creative solutions. The requirement of an adequate stimulus that induces NP biosynthesis brought forth overcoming strategies that e.g. mimic the suitable environmental condition in the lab. Upon success, this eventually leads to expression of dormant biosynthetic genes and in consequence triggers NP production.
 Fraunhofer Institute for Molecular Biology and Applied Ecology IME
Fraunhofer Institute for Molecular Biology and Applied Ecology IME